Storing data is a simple task isn’t it? Memory is relatively cheap and after all data wants to be free doesn’t it? How hard can it be? Here are my ten tips for building a terrible database.
TIP ONE Make submission difficult
Scientists are smart people so there’s no need to bother wasting time and money on usability issues. Eventually they’ll figure out how to get it right and at least some of the important information will be submitted. Who cares if everyone submits to a rival database, just because it’s easier to use?
TIP TWO Have a support service that is available 9-5 Mon to Fri GMT
After all scientists are renown for working 9-5 and the only science that matters is in Europe… isn’t it?
TIP THREE Don’t let your file formats interconvert
Under no circumstances should your data from one piece of equipment in a specific file format be converted into a common and searchable one, or even be read without proprietary software. Particularly, ignore the pioneering work of the open microscopy environment: format standardisation is for wimps.
TIP FOUR Keep your database independent
Stand out from the crowd by ensuring your data do not link to other databases. Who wants their data to be found via a sequence search on GenBank or through links from UniProt? Data wants to be free but it doesn’t necessarily want to be found.
TIP FIVE Totally trust your automated systems
Books can be ordered on Amazon without any manual intervention so why would it be needed on a database? Most of the well known biological databases have curators who check the submissions, ensuring that they are complete and accurate as far as possible. What a waste of money – nobody minds incomplete data sets, missing experimental conditions etc.
TIP SIX Do not provide a permanent, unique identifier
The PDB uses identities (e.g. 1ubq) and the Gene Expression Ominibus uses an accession number – as do many other databases, but this looks like another hassle you don’t need. We all need a good place to bury bad data.
TIP SEVEN Make sure reviewers can’t see raw data
Don’t devise a simple way for journal reviewers to check data that is part of paper going through peer review. Reviewers LOVE to receive emails with thousands of huge images attached.
 TIP EIGHT Include a 44-page getting started guide
TIP EIGHT Include a 44-page getting started guide
Scientist have lots of spare time and are very keen to read through a 44-page quick-start guide to your database because you’ve followed tip one and ensured that the database is very difficult to use. Even better, provide at least a 50-page guide for reviewers. The only people less busy than your submitters are the reviewers. It’s a well known fact.
TIP NINE If you include a search option, make sure it only works in UK English
Or in US English, but certainly not in both. People foolish enough to search for crystallization and not crystallisation don’t deserve to find anything in a database.
TIP TEN Do not develop good visualisation tools
Scientists love data. Pages and pages and pages of it. Making it simple to see connections between different datasets would just make it too simple. Scientist love a challenge.


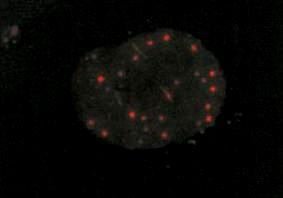
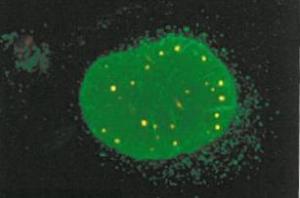
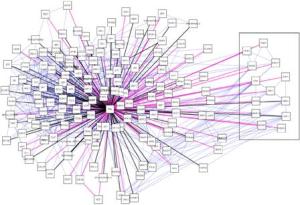
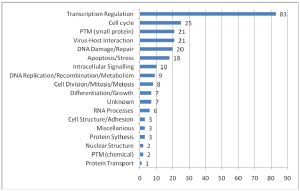
 I’ve been to a few conferences recently and I’ve witnessed a divide opening up between the scientists that use high-throughput methods and everybody else. This I think is partly because although large datasets look impressive, we’re just not sure what it all means yet. Some researchers have even said to me that the interest of some of the top journals in publishing large datasets is simply because they lead to good citations, and help the impact factor. A recent paper on the ‘PML interactome’, which I describe below, is a nice example of how assembling the data in one place gives a very good overview of the situation and provides some functional clues too.
I’ve been to a few conferences recently and I’ve witnessed a divide opening up between the scientists that use high-throughput methods and everybody else. This I think is partly because although large datasets look impressive, we’re just not sure what it all means yet. Some researchers have even said to me that the interest of some of the top journals in publishing large datasets is simply because they lead to good citations, and help the impact factor. A recent paper on the ‘PML interactome’, which I describe below, is a nice example of how assembling the data in one place gives a very good overview of the situation and provides some functional clues too.